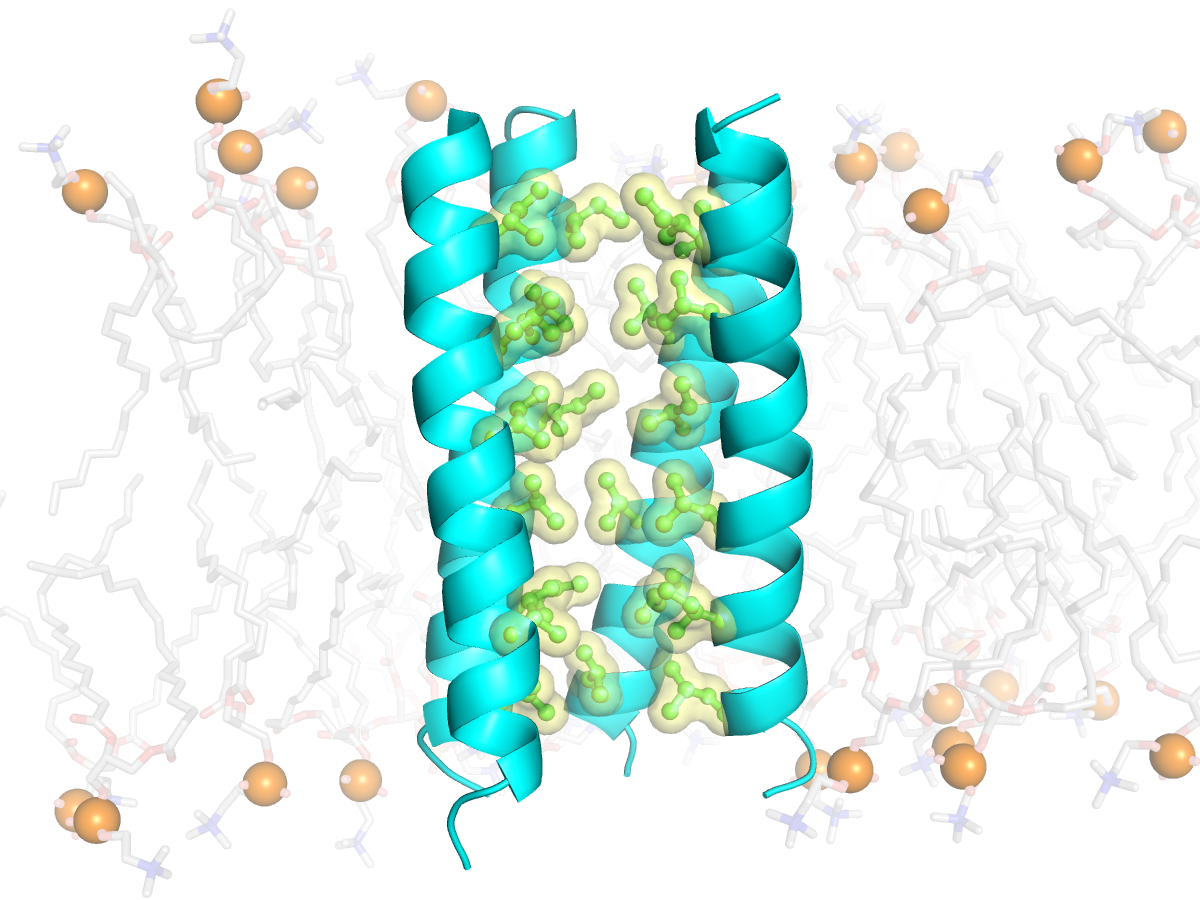
Membrane protein design & molecular biophysics
We pair biophysical theory (energy calculations + atomic simulations) with informatic mining of molecular databases to derive data-driven hypotheses for how membrane proteins fold. This engineering approach allows us to stabilize new custom membrane proteins and novel lipid-embedded protein-protein interaction partners from scratch.
We use computational design alongside protein total synthesis, NMR, crystallography, and biophysical techniques to challenge our understanding of membrane protein physical chemistry and molecular events in disease.
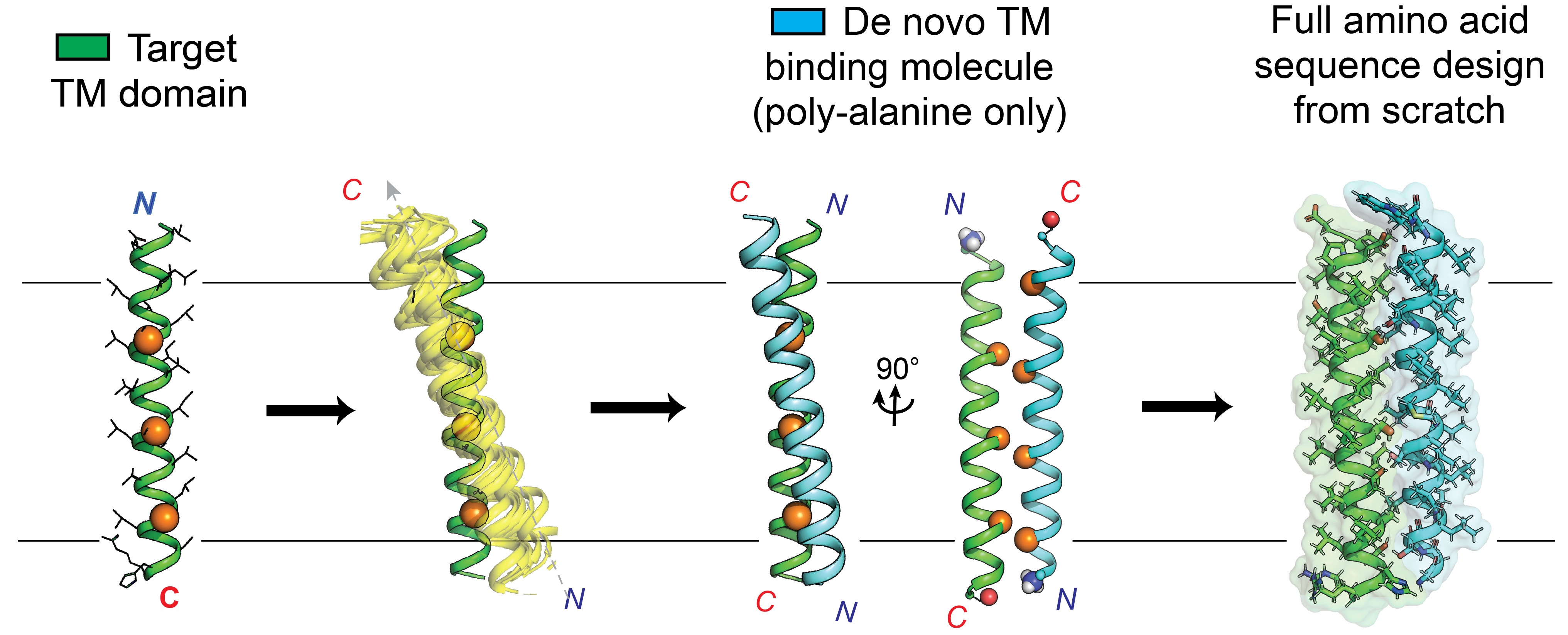
Targeting membrane proteins directly by their lipid-embedded domains: Transmembrane “antibody” miniproteins
We develop and apply computational algorithms to design small lipophilic proteins from scratch that bind proteins directly at their transmembrane domains to alter the structural and functional states. These custom chemical probes allow us to dissect the active role TM domains play in signaling and transducing protein conformational changes as well as stabling critical states for structural studies (X-ray/EM). Thus, membrane proteins without considerable folded water-soluble domains can still be targeted at their ‘achilles heel’ TM domains.
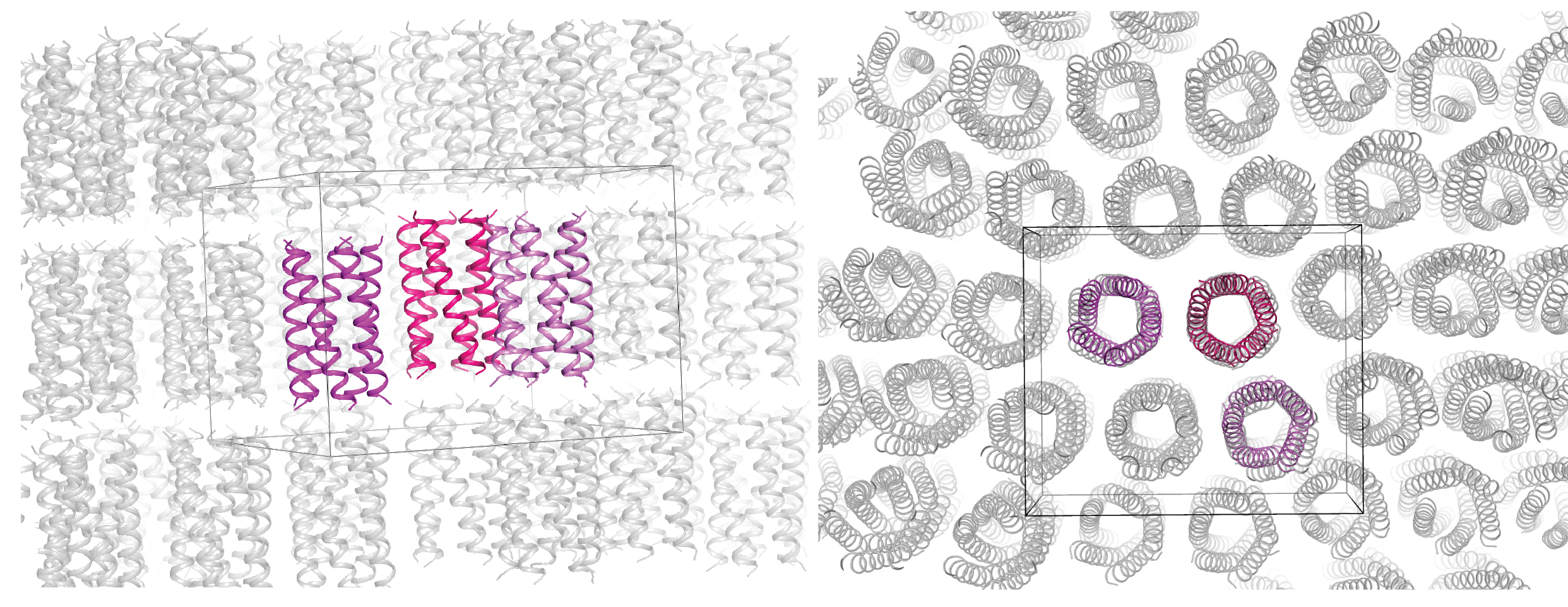
Design of simple synthetic model membrane proteins to directly test fundamental biophysical questions
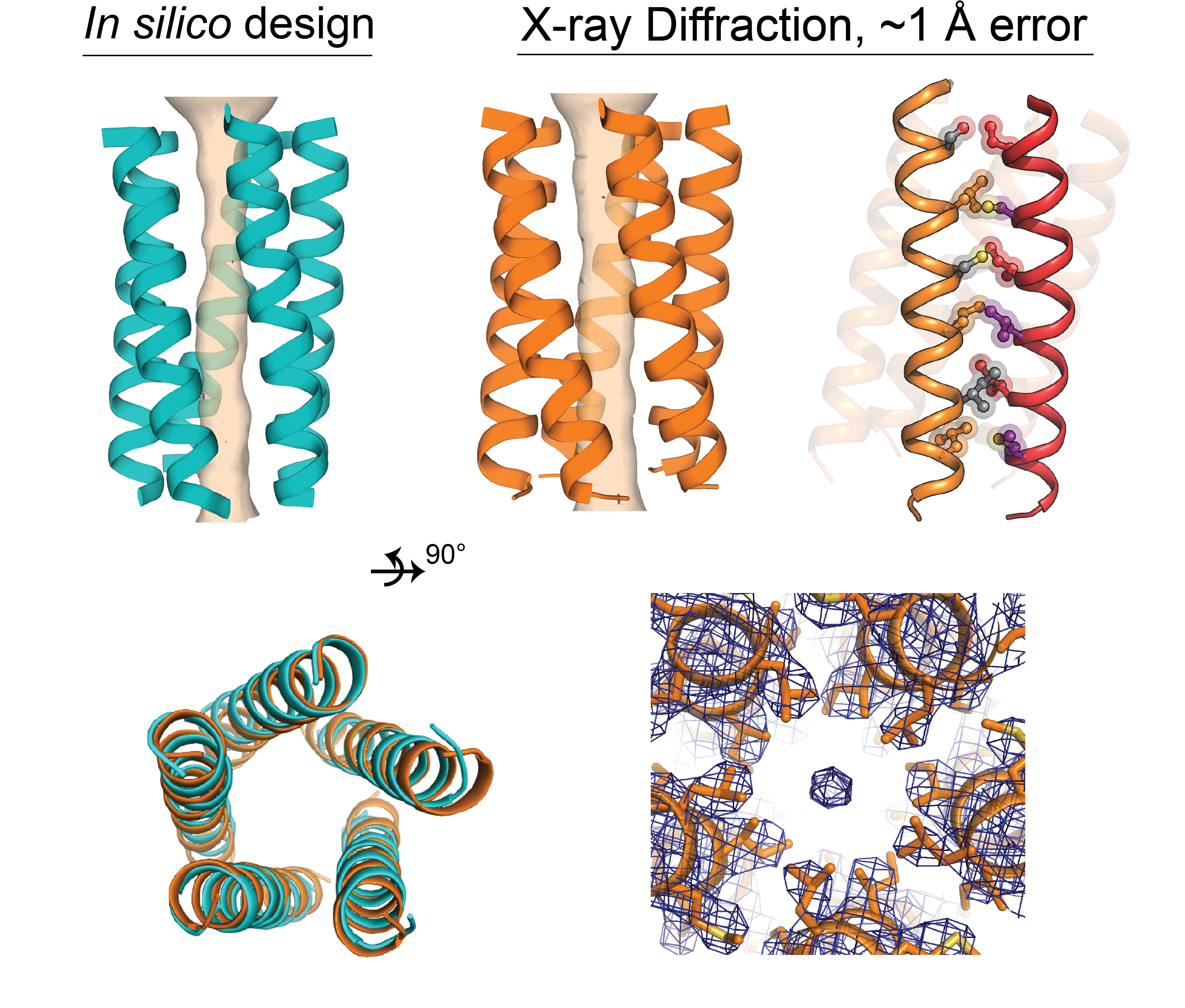 We are interested in the design principles for constructing membrane proteins, which we then apply to engineering custom synthetic proteins for biotechnology applications or to understand natural proteins in disease. Classically, biophysics & biochemistry researchers try to assess general theories by studying specific well-known model membrane proteins which are often large, complex, and poorly behaved (a PhD student’s nightmare) thus can cloud biochemical interpretation. We instead use protein design as an approach to construct small simple synthetic membrane proteins as easy-to-study model proteins, custom made to ask direct and basic and generalized ‘textbook’ questions concerning protein physical chemistry in the unfamiliar non-aqueous environment of lipid bilayers.
We are interested in the design principles for constructing membrane proteins, which we then apply to engineering custom synthetic proteins for biotechnology applications or to understand natural proteins in disease. Classically, biophysics & biochemistry researchers try to assess general theories by studying specific well-known model membrane proteins which are often large, complex, and poorly behaved (a PhD student’s nightmare) thus can cloud biochemical interpretation. We instead use protein design as an approach to construct small simple synthetic membrane proteins as easy-to-study model proteins, custom made to ask direct and basic and generalized ‘textbook’ questions concerning protein physical chemistry in the unfamiliar non-aqueous environment of lipid bilayers.
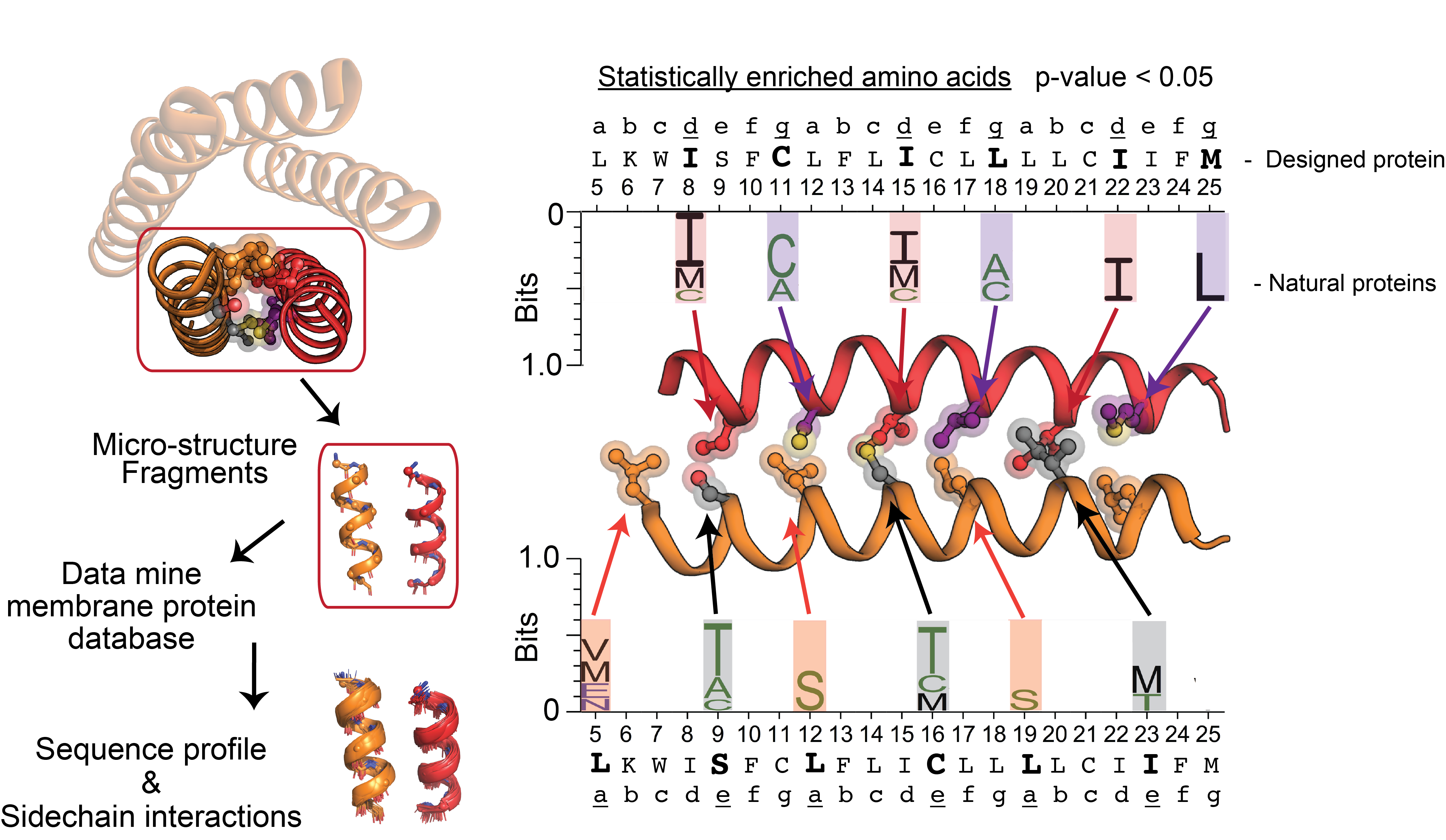
Bioinformatic data mining & molecular dynamics simulations of membrane proteins
 We mine databases of biomolecular structures to reveal trends of how protein atomic groups preferrentially interact, informing our design of new functional and stably folded proteins. Our groups is focused on computational approaches for integrating sequence-structure relationships into atomic models for both synthetic and natural membrane proteins. We pioneer approaches for integrating diverse forms of biochemical data into atomic protein models and molecular dynamics simulations to reveal relevant protein conformations, stabilizing motifs, and dynamics instrumental to stucture, function, and evolution - typically within the hydrophobic water-devoid lipid membrane milieu.
We mine databases of biomolecular structures to reveal trends of how protein atomic groups preferrentially interact, informing our design of new functional and stably folded proteins. Our groups is focused on computational approaches for integrating sequence-structure relationships into atomic models for both synthetic and natural membrane proteins. We pioneer approaches for integrating diverse forms of biochemical data into atomic protein models and molecular dynamics simulations to reveal relevant protein conformations, stabilizing motifs, and dynamics instrumental to stucture, function, and evolution - typically within the hydrophobic water-devoid lipid membrane milieu.